Disambiguating Medical Terms using MedSpacy Natural Language Processing
Learn how to use the on_match function in MedSpacy to add disambiguation logic to your named entity recognition (NER) rules.

Learn how to use the on_match function in MedSpacy to add disambiguation logic to your named entity recognition (NER) rules.
Introduction
MedSpacy is a rule based NLP library built on top of Spacy for Python. It functions very similarly to Spacy, but with custom token splitting and some other tweaks to optimize for processing of medical texts.
I'm using MedSpacy to extract data from unstructured text within our electronic health record. It runs locally, so no patient data leaves my institution, and as a rule-based solution I have a high level of control over how it functions compared to something like a large language model.
The overall process involves writing rules to identify words or phrases, tagging and applying labels to them, and extracting that data to a structured format.
Unfortunately, many terms and acronyms in medicine are ambiguous without knowing the context in which they're used. This makes it challenging for rule based algorithms to properly identify terms and phrases without knowledge of the surrounding text.
Lucky for us, the on_match function in MedSpacy lets you write your own disambiguation rules to solve this problem.
How Does On-Match Work?
With rule based tagging, a word, phrase, or pattern (generally referred to as an "entity") is recognized in text. In MedSpacy, these entities are typically some sort of medical problem or treatment.
Your rules can have additional custom attributes to be included when a named entity is tagged. These could be ICD codes, more specifics about the diagnoses or treatments, or anything else you need to accomplish your NLP task.
Typically, the matching rules will define how an entity will be labeled. Once it is matched, the labels are applied, and the pipeline moves on.
On-Match allows you to add another layer of logic or processing to be run when a certain entity is identified. This gives you greater control over how labels are applied to entities, and can be used to disambiguate medical terms based on context.
Today we'll be writing a Python script with MedSpacy that identifies the phrase "Stage IV" (and its variations) and attempts to disambiguate what type of disease it is referring to before assigning a label.
Let's look at the code. Here is a Colab notebook with the full code if you want to jump right into it yourself.
Installing Dependencies
pip install medspacyRunning this will install all necessary packages and dependencies, including the right version of Spacy.
For this particular project, you will need to import the following packages:
import medspacy
import spacy
from spacy.tokens import Span
from medspacy.ner import TargetRule
from medspacy.target_matcher import TargetMatcher
from medspacy.visualization import visualize_ent, visualize_depSetting Up Your MedSpacy Pipeline
Spacy and MedSpacy both allow you to build out a pipeline for your natural language processing. Depending on what you are trying to accomplish, you can mix and match various components. These include pre- and post- processing components, as well as advanced NLP components like transformers, LLMs, and more.
MedSpacy adds four new pipeline components to be thrown into the mix:
medspacy_tokenizer: (Automatically included in your pipeline) Custom tokenizer for clinical text.medspacy_pyrush: Clinical sentence splittermedspacy_target_matcher: Extended rule-based matchingmedspacy_context: Customizable contextual analyzer
To set up your pipeline, run the following:
nlp = medspacy.load()To see what is currently included in your pipeline:
nlp.pipe_names
#['medspacy_pyrush', 'medspacy_target_matcher', 'medspacy_context']By default the tokenizer (not shown in pipe_names), sentence splitter, and context analyzer are included with the target matcher. You can remove these if you want, but they won't get in the way of our project in any way.
Setting Up Target Rules and Custom Attributes:
To set a basic Target Rule, you call TargetRule and provide (literal, label). literal is what the rule will match, and label is what it will mark it as. In non-medical NLP, labels could be something like person, organization, event, date, etc. In medical text, often it is something like diagnosis, treatment, or something similar.
The most basic rule looks like this:
target_rules_basic = [
TargetRule("Stage IV", "Cancer")
]This will only apply to exact matches, including capitalization, formatting, punctuation, etc.
If you add a pattern to your rule, this will be matched instead of literal, and allows for you to use a list of dictionaries or regular expressions for your match.
Using a pattern, our target rule for the variations of "Stage 4" that are frequently written in medical text would look like the following:
target_rules = [
TargetRule("stage_4", "Problem",
pattern=[
{"LOWER": {"IN": ["stage"]}},
{"IS_PUNCT": True, "OP": "?"},
{"LOWER": {"IN": ["iv","4","four"]}},
],
attributes={"problem_type": "unknown"}
)
]The complexities of the MedSpacy target matcher could be a whole post in itself, but this will capture all the (correctly spelled) variations of "Stage 4" and add a problem_type attribute of "unknown".
In order to use custom attributes ("problem_type" in this case,) you need to create the attribute first. I usually include the following line of code with my library imports to get it out of the way. You can give the attribute any name you want, and add multiple attributes for complex NLP tasks.
Span.set_extension("problem_type", default="")The on_match Function
Spacy and MedSpacy have the ability to call an additional function whenever a certain named entity is matched.
Here's a function which will disambiguate "Stage IV" as either cancer, wound, or pulmonary disease. This function looks at the words surrounding the matched "stage 4" for key words that provide additional context and assigns the problem_type based on that. If none of the key words are matched, it returns "unknown."
def disambiguate_stage_4(matcher, doc, i, matches):
match_id, start, end = matches[i] #sets span variables based on the matched entity
ent = doc[start:end] #Defines ent based on the start + end of the matched entity within the target doc
surrounding_tokens = doc[start-1:end+3] #defines surrounding tokens
#looks for specific words in the surrounding tokens to help disambiguate the matched entity
if any(tok.lower_ in ["cancer","malignant"] for tok in surrounding_tokens):
ent._.set("problem_type", "cancer")
print(f"Updated {ent.text} to problem_type: cancer")
elif any(tok.lower_ in ["sacral","wound"] for tok in surrounding_tokens):
ent._.set("problem_type", "wound")
print(f"Updated {ent.text} to problem_type: wound")
elif any(tok.lower_ in ["sarcoid","sarcoidosis","copd","gold"] for tok in surrounding_tokens):
ent._.set("problem_type", "copd")
print(f"Updated {ent.text} to problem_type: wound")
#If none of the above are matched, sets problem type to unknown
else:
ent._.set("problem_type", "unknown")
print(f"Updated {ent.text} to problem_type: unknown")Now, we can add this to our target rule.
target_rules = [
TargetRule("stage_4", "UNKNOWN",
pattern=[
{"LOWER": {"IN": ["stage"]}},
{"IS_PUNCT": True, "OP": "?"},
{"LOWER": {"IN": ["iv","4","four"]}},
],
on_match=disambiguate_stage_4)
]When the term "Stage 4" or its equivalents, as defined by our pattern, is matched, instead of simply tagging the entity and moving on, it will run the disambiguate_stage_4 function to search surrounding tokens for additional context that helps clarify what is being referenced.
Lastly, we need to add the target_rule to our target_matcher so that it is included when we run our NLP pipeline:
target_matcher = nlp.get_pipe("medspacy_target_matcher")
target_matcher.add(target_rules)Running the Pipeline
To run text through the pipeline, we define the text, and then pass it through our pipeline using nlp().
text1 = 'gentleman with stage IV colon cancer, bacteremia, also newly developed stage IV sacral wound.'
text2 = 'gentleman with stage IV sarcoidosis'
text3 = 'stage 4 copd'
doc1 = nlp(text1)
doc2 = nlp(text2)
doc3 = nlp(text3)
#Updated stage IV to problem_type: cancer
#Updated stage IV to problem_type: wound
#Updated stage IV to problem_type: pulmonary
#Updated stage 4 to problem_type: pulmonaryWe can run the following to visualize the docs and extract the label and problem_type attribute:
for doc in [doc1, doc2, doc3]:
visualize_ent(doc)
print('=' * 50)
for ent in doc.ents:
print(ent.text,' | ', ent.label_,' | ',"Problem Type:", ent._.problem_type)
print('=' * 50)
print('')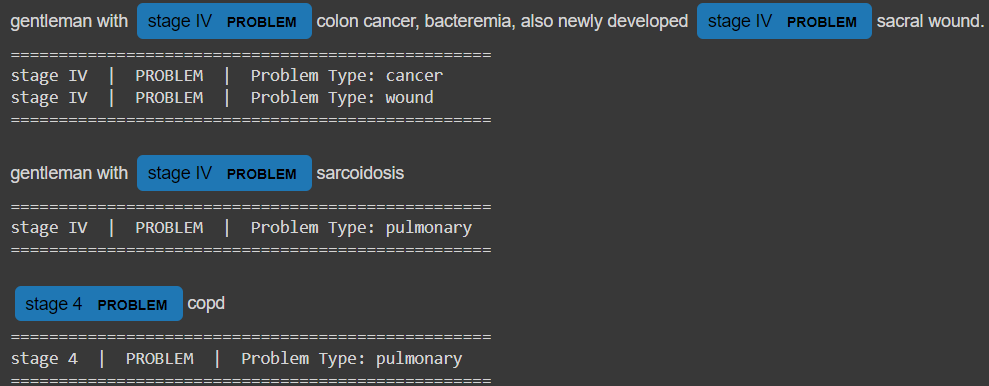
As you can see, all of the instances of "stage IV" were matched and labeled as problems, with different problem_types based on their surrounding text.
The functions that are called on_match aren't limited to those like the above example. For example, you could write a function which calls an LLM API when a certain phrase is matched to help perform more complex disambiguation or other tasks on the relevant text.
Conclusion
In this post, you have learned:
- What MedSpacy is and how to download and install it
- How to initiate an NLP pipeline
- How to set up target rules
- How to add those rules to your NLP pipeline.
- How to use the
on_matchmethod to augment the matching logic to disambiguate medical terms.
Here is the Colab notebook again. Play around with the rules, on_match function, and example texts and see what you can come up with.
It took me a while to piece together exactly how to use on_match. Hopefully this post saves you a bit of time as you're exploring MedSpacy and working on your own NLP projects.
MedSpacy has a whole host of tools that can be used for medical NLP. Since my primary project right now is an NLP project, I'll be writing about it for the foreseeable future. If there is anything specific you want to learn about, let me know.
Additional Resources
I'm self-taught and still learning on a daily basis. Writing is a way for me to solidify my knowledge and share what I have learned. As a student, I will also make mistakes. If you see something I'm doing wrong, or could be doing better, please let me know.